Thanks to the efforts of geneticists the story of the extinction of the Spanish Habsburgs is now well known. They are in short a case study in the disastrous consequences of an inbred pedigree. The downsides of inbreeding are to some extent intuitively understood by all, especially consanguineous relations between first order relatives. Though I'm willing to bet that all things equal inbred individuals are not as attractive or intelligent as outbred individuals, the literature in this area for humans is surprisingly thin. A major problem is controlling for confounds; all things are often not equal (e.g., imagine if inbreeding is more common in marginal isolated communities, which is often true in the West. See Consanguinity, Inbreeding, and Genetic Drift in Italy, where it is obvious that the less developed areas of Italy had elevated rates of marriage between relatives despite Catholic discouragement of the practice). But the case that inbreeding results in the expression of deleterious recessive diseases is more straightforward. The rarer the disease, the higher the proportion of individuals who are affected who are the consequence of inbreeding. This is due to the logical fact that very rare alleles tend not to come back together in homozygote form due to the character of the Hardy-Weinberg equilibrium. If the recessive trait is caused by a minor allele with a frequency of p, p2 can converge upon zero very rapidly as p decreases in frequency. At p = 0.1 the recessive trait will express in 1% of the population (so p/p2 = 10). At p = 0.01 the recessive trait will express in 0.01% of the population (so p/p2= 100). And so forth.
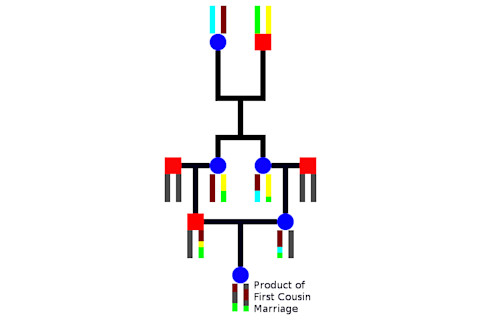
Inbreeding changes the equation, so to speak, because you are no longer dealing with random breeding. Instead you are bringing together individuals with the same rare complement of deleterious alleles which may exhibit recessive expression. To make this concrete I drew up a simple stylized pedigree where I trace the descent of one pair of chromosomes from the generation of two grandparents down to the offspring who is the product of a first cousin marriage, and so share those same two grandparents. I have illustrated the process of recombination, where two homologs break apart and are recombined to produce a synthetic new sequence. What you see is that by the end of the process some segments from one of the four chromosomes of the shared grandparents have been passed down so that they produce two very long runs of homozyogsity (at least on the genomic scale).* This is problematic. If there are any recessive deleterious mutations along this segment then you by definition have a homozygote at that position (barring a rare back mutation). Obviously individuals who have recent ancestors who appear in multiple positions in their pedigree have a much higher likelihood of having homozygous genotypes, which are often not optimal. In the more extreme cases this produces recessive diseases (and because inbreeding reduces effective population and increases the power of random genetic drift deleterious recessives can even fix within a population rather quickly). Even in cases where individuals are healthy there is likely a fitness drag due to elevated homozygosity across the genome.
And this problem varies quite a bit by locale. Some populations are much more inbred than others, because of cultural practices. In the Arab Muslim world marriages between the offspring of brothers are a common way to cement relations between branches of a lineage. In some South Indian castes marriages between uncles and nieces are common, while in other cases marriages between the offspring of a brother and a sister are preferred. Finally, even though familial exogamy is enforced among many North Indian Hindus, a population bottleneck during the founding of the groups, and subsequent intra-caste endogamy, have resulted in a likely higher load of recessive diseases among these populations, despite their aversion to consanguinity.
But there is hope. Generations of inbreeding, resulting in near pedigree collapse, can be abolished by one generation of outbreeding! How is this possible? Think back to the example of the chromosome with long homozygous stretches. Imagine a magical individual who is 100% homozygous, but alive and fertile. Likely they are a repulsive nincompoop due to the enormous disease burden, but in this fanciful world they find someone who mates with them who is not related to them. Most of the deleterious recessive traits will immediately become masked in the offspring, because at the locations where the inbred individual carries functionally defective alleles their mate will carry the wild type variant.
What does this mean in the "news you can use" department? If your partner comes from an inbred lineage then don't worry about your offspring (at least for genetic reasons). The outbreeding event will result in a genotype unburdened by deleterious recessive expression. People are always looking for ways to be socially productive and altruistic, and marrying someone from an inbred lineage is one way to guarantee that the world is filled with healthier and more beautiful people.
* A friend who is the product of a first cousin marriage has many 30-50 megabase homozygote segments.